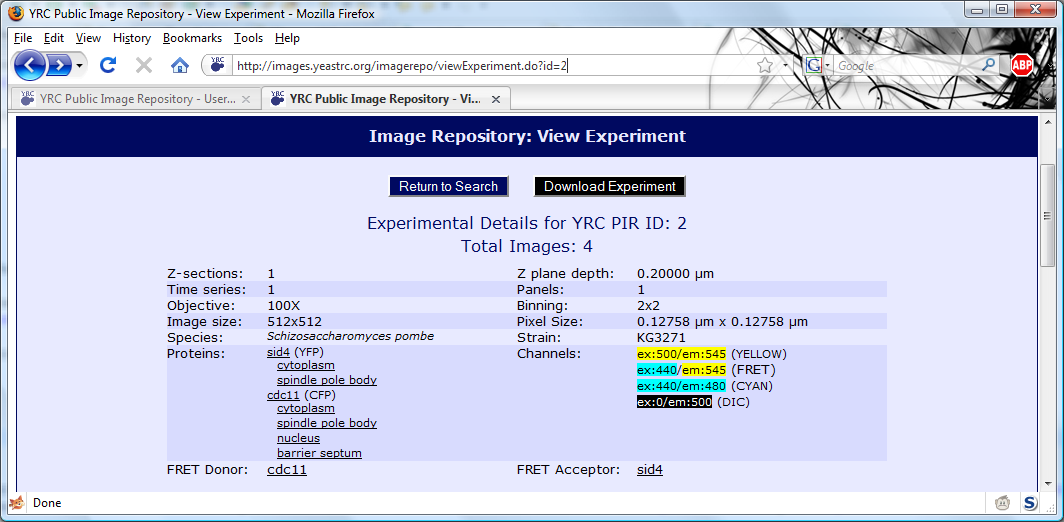
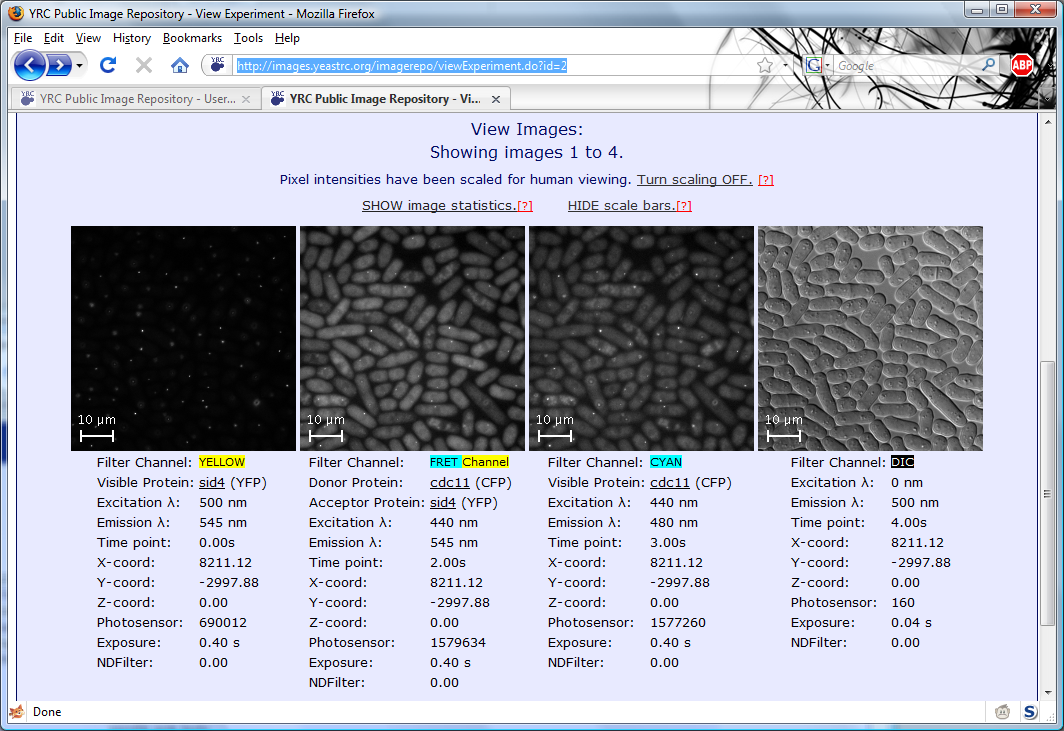
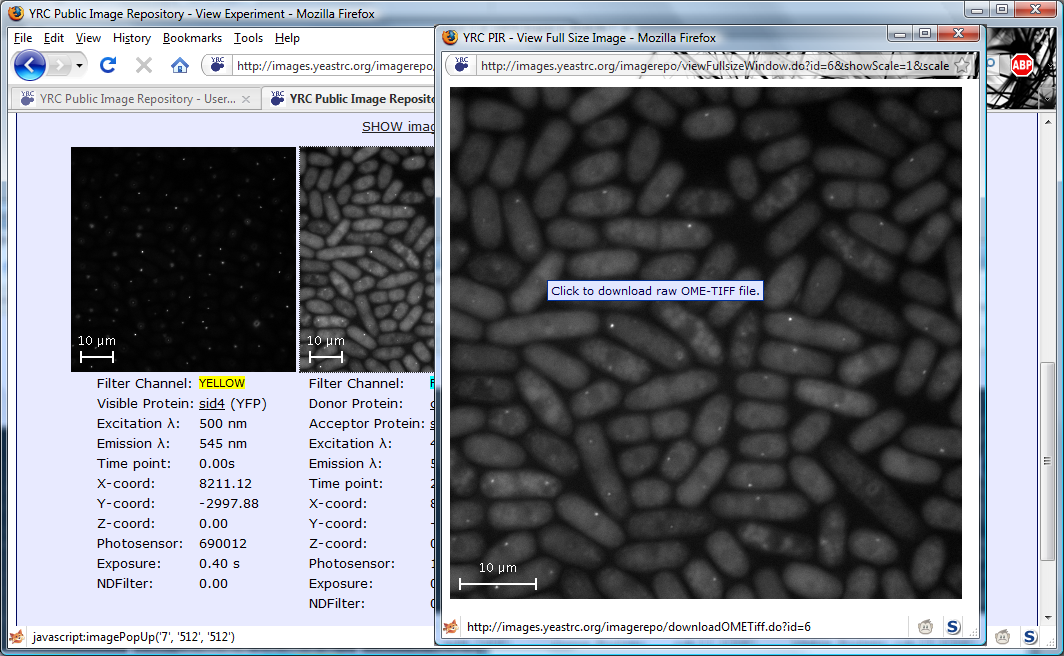
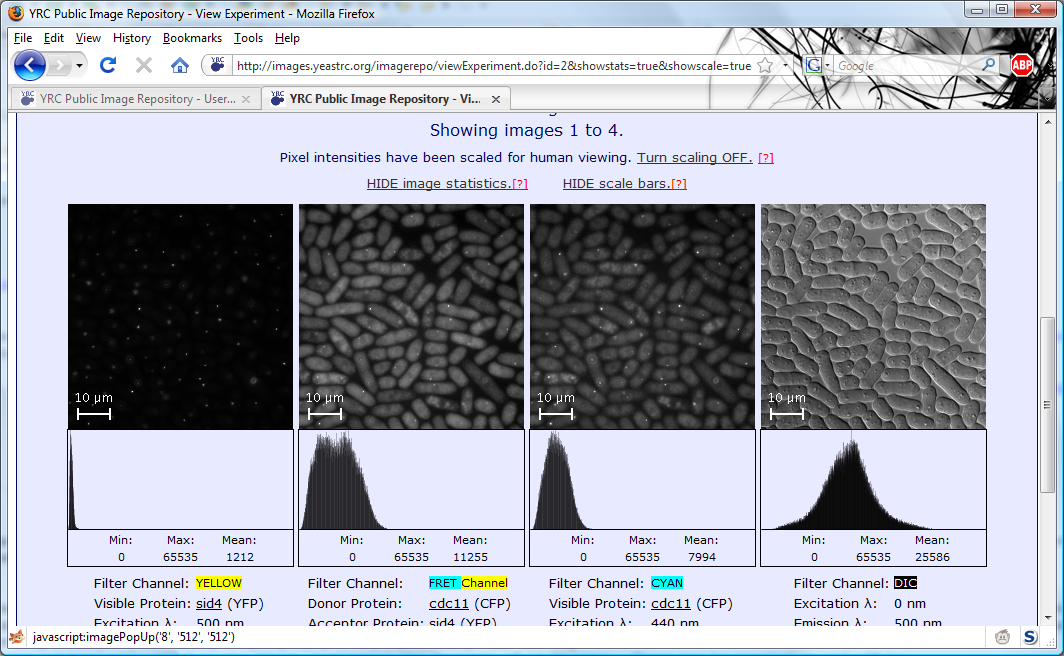
All searches in the YRC PIR begin with the same search form. This form is the default starting page for the site,
and may be reached at any time by clicking the "Search" tab at the top of every page.
The form is used to define parameters for searching the experiments in the database. All matching experiments
will be returned to the user on a search results page (described later). Note: By default, not specifying any
additional parameters on the search form will return all results in the database.
The following is a description of the fields present on the search form:
i. Data source
Only experiments from the selected source of data will be included in the results.
ii. Include FRET experiments?
Should FRET experiments be included in the results? For information about FRET, please see the
YRC FRET tutorial.
- No - FRET experiments will not be included
- Yes (Inclusively) - FRET experiments are included in results (in addition to non-FRET experiments)
- Yes (Exclusively) - Only FRET experiments will be included
iii. Number of channels
Only experiments that contain the specified number of
channels will be included in the search results.
iv. Number of Z-sections
Only experiments that contain the specified number of
Z-sections (number of focal planes) will be included in the search results.
v. Number of time series
Only experiments that contain the specified number of
time series will be included in the search results. Time series are defined as
the number of times the each channel was imaged at the same X and Y coordinates.
vi. Objective
The magnification power of the objective used to image the experiment.
vii. Binning
The binning that was applied to the image when it was captured by the camera.
See the FAQ.
viii. Image size
The dimensions, in pixels, for the images captured in the experiment.
ix. Select protein
All matching experiments will include the protein selected by this option. To add a protein to the search, click the
[Add Protein] link. A new window
will appear allowing you to search for the protein that you wish to include in your image search:
Entering the name or partial description of the protein will yield search results:
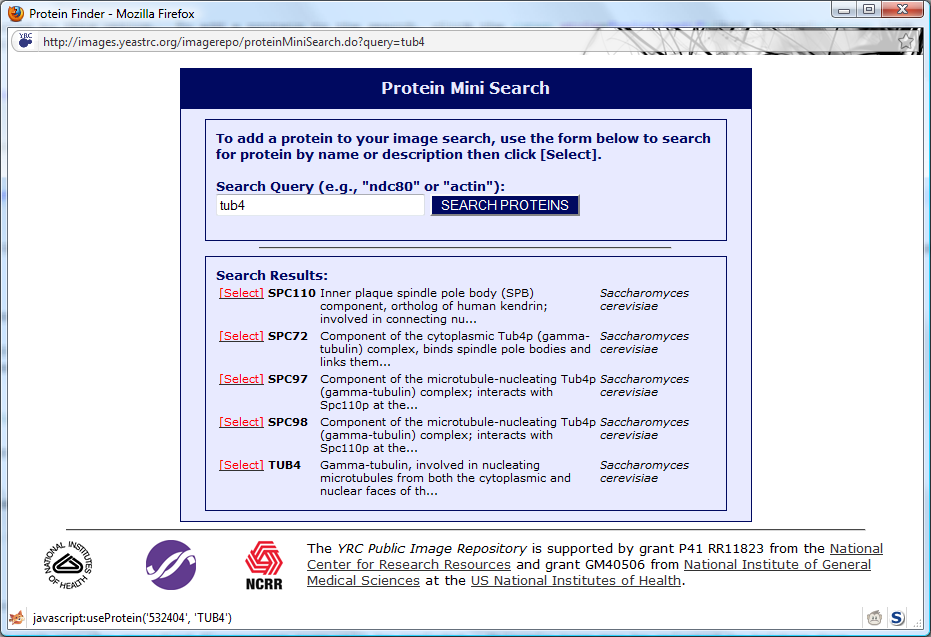
Once you have located your protein of interest, click the
[Select] link next to the protein name, and that protein will be added to your search (and the search
window will automatically close).
x. Select second protein
All matching experiments will include both this protein and the previously selected protein. This is provided as a means of finding colocalization experiments
and FRET experiments containing specific pairs of proteins. The process of adding a second protein works exactly the same as the first protein, as described above.
xi. Species
All matching experiments will contain proteins from the selected species.
xii. Strain name
Only experiments imaging cells of the specified strain will be returned. These are strings used internally in the YRC PIR and are displayed in search results and
in experiment view pages.
xiii. Limit to following fluorescent tags
Only experiments containing at least one protein tagged with one the specified flourescent tags will be included. Multiple tags may be selected by holding down
control (in Windows or Linux) or command (on a Macintosh).
xiv. Gene Ontology
You may specify one or more Gene Ontology terms and all experiments returned will contain at least one protein annotated with any of the specified terms. To specify a term, click
the
[Use GO Searcher] link. A window will appear allowing you to search the Gene Ontology:
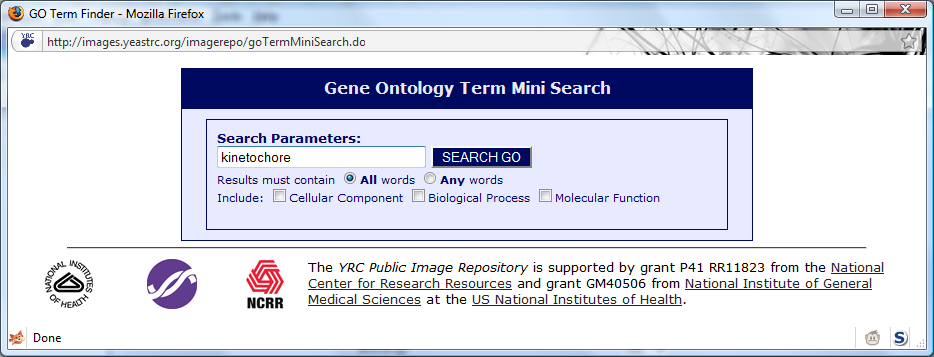
In the example, kinetochore was searched for. The search results appear as:
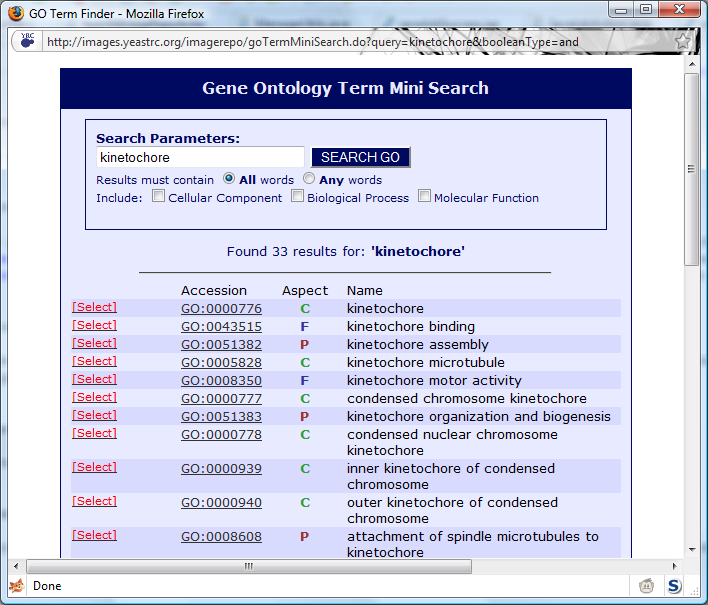
Clicking
[Select] next to the desired GO term will add that GO term to the Image Repository search.
To add additional GO terms to the search, click the +Add Another GO Term link and repeat the above procedure. You may add up to 5 GO terms to your search.
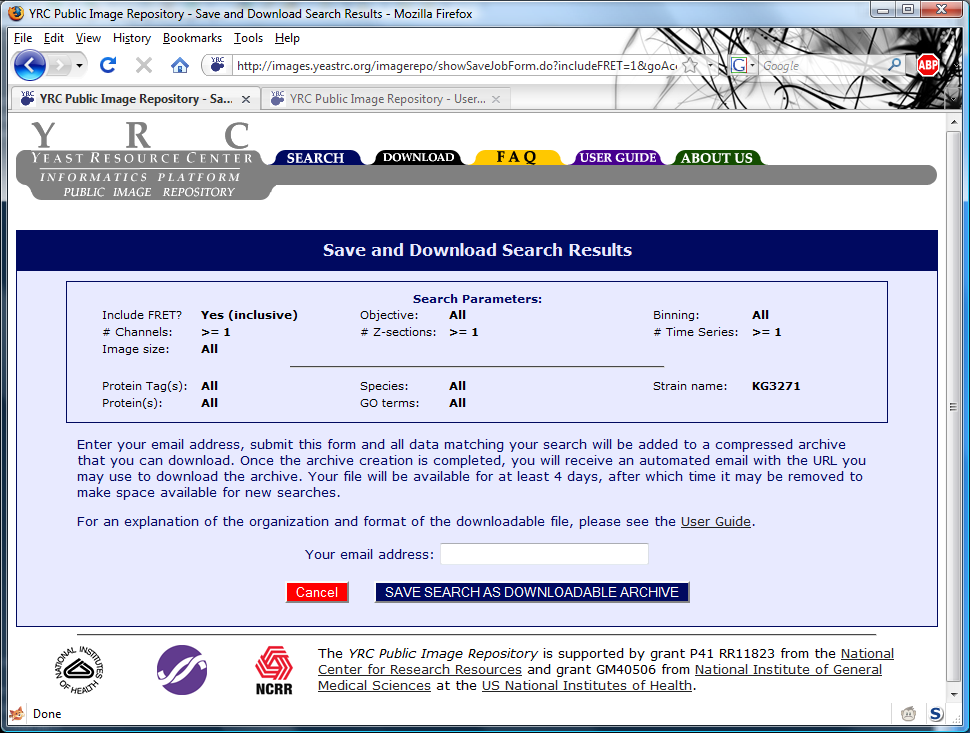
An screen shot of sample search results:
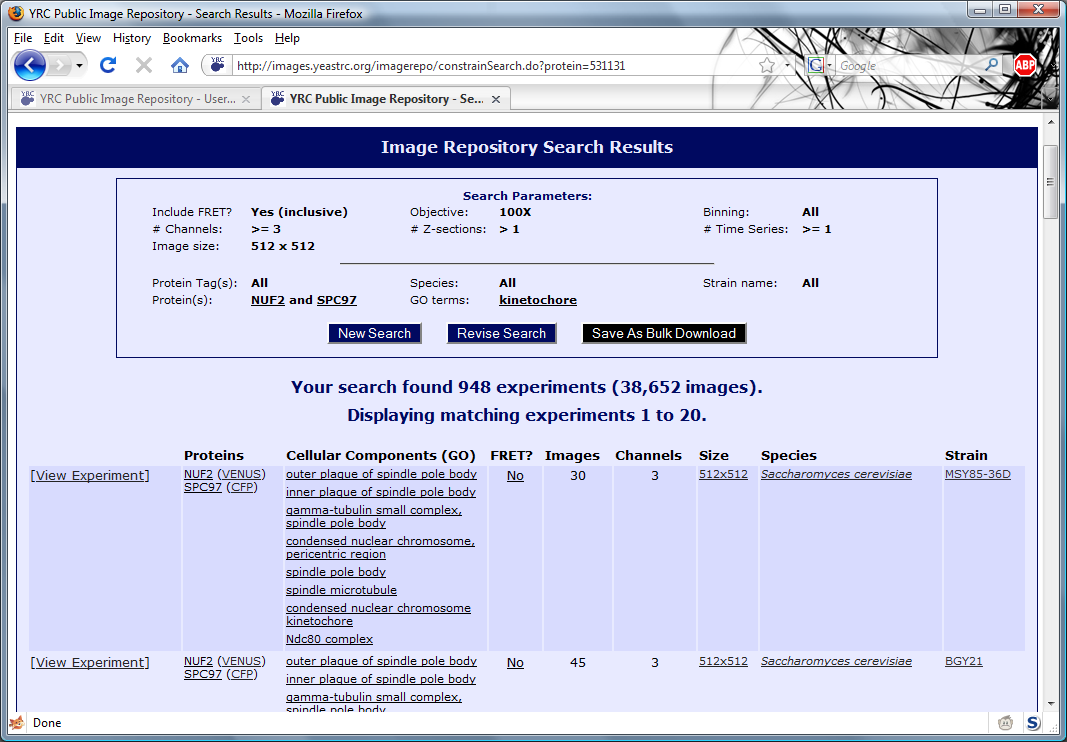
Listed in the box at the top are the search parameters corresponding to the listed search results. The "New Search" button will return the user to a default search form. The "Revise Search"
button will return the user to the search form that is filled in with current search parameters. The "Save as Bulk Download" button allows the user to save all data matching the current search
as a single downloadable archive. This is described in more detail in the "Downloading Data" section.
Following the search parameters list are the search results, themselves. They are listed 20 per page, and may be browsed by clicking "Previous 20 Results" or "Next 20 Results" at the bottom of
the search results. Clicking "[View Experiment]" will take the user to the experiment view page where images and metadata may be viewed. The columns contained in the search results are
described in more detail in
the FAQ.
In addition to clicking the "Revise Search" button under the search parameters, the links present in the search results may be used to refine the search. With the exception of proteins, GO terms
and whether or not to include FRET experiments, simply clicking links present in the search results will add that constraint to your search. For example, clicking on "BGY21" in the "Strain"
column in the example results above will add that constraint to the search and automatically research the database.
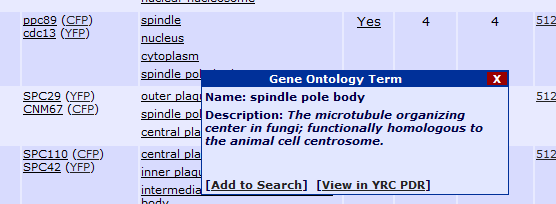
In the case of protein names and GO terms in the results, placing your mouse arrow over a protein or GO term will produce the following tool tips to appear:
GO term:
 Protein:
Protein:
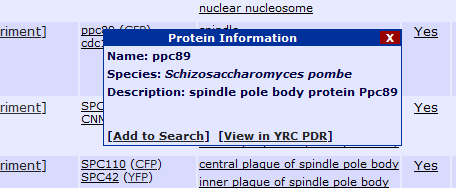
Clicking "Add to Search" will add this protein or GO term as a constraint to the current search. If the search already contains a protein, this protein will be added as
a second protein and all results will need to contain both proteins. If the search already contains two proteins, this new protein will replace the second protein in
the search constraints. Clicking "View in YRC PDR" will open a page from the YRC Public Data Repository containing more detailed information about the protein or GO term.
In the case of FRET, placing your mouse arrow over the FRET column yields the following tool tip:
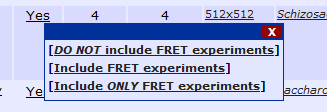
Clicking any of the links will add the corresponding constraint to the current search, replacing the current FRET constraint.